Antisense oligonucleotides
Antisense oligonucleotides (ASOs) are oligonucleotides that induce gene silencing. Typically, ASOs are 15–22 nucleotides in length and are designed to bind complementary RNA targets, ultimately facilitating their degradation via the RNase H pathway. Our well-established expertise in oligo synthesis makes us a trusted vendor for ASOs.
Ordering
- Choose the scale, modification, and purification options to meet your knockdown experiment needs.
- Increase intracellular stability and reduce toxicity with nuclease-resistant modifications and flexible chimeric designs.
- Achieve effective gene silencing for your targets in cellulo.
Antisense oligonucleotides
We offer chemically synthesized and modified antisense oligonucleotides (ASOs) at different scales with different purification options in tube or plate formats. ASOs are ideal for inhibiting gene expression in vitro, in cellulo, and in vivo studies. Affinity Plus™ ASOs incorporate locked nucleic acids for enhanced stability and nuclease resistance. HPLC-purified ASOs include sodium salt exchange.
Custom ASOs can include non-standard lengths or modifications, as well as, designs for splice switching or steric blocking.
Don't see what you are looking for? We are continually expanding our products, and we may have what you need. For assistance, Contact us
Product details
Antisense oligonucleotides (ASOs) are DNA oligos, typically 15–22 bases long designed in antisense orientation to the RNA of interest. Hybridization of the ASO to the target RNA triggers RNase H cleavage of the RNA, which can inhibit the function of non-coding RNAs (e.g., miRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and lncRNAs) or prevent protein translation of mRNAs. To increase nuclease resistance, we recommend adding phosphorothioate (PS) modifications to the oligo. In the IDT ordering system, use an asterisk to indicate the position of a phosphorothioate internucleoside linkage. Also consider adding modified bases, such as 2′-O-methyl (2′-OMe) or Affinity Plus™ locked nucleic acid bases, in chimeric antisense designs to increase nuclease stability and affinity (Tm) of the antisense oligo to the target mRNA [1]. Substitution of 5-methyl dC for dC in CpG motifs will slightly increase the Tm of the antisense oligo.
| Examples of RNase H active antisense oligos | |
|---|---|
| Sequence (with modification) | Modifcation description |
| 5′ A*C*G*C*G*C*G*A*C*T*A*T*A*C*G*C*G*C*C*T 3′ | DNA, All PS |
| 5′ /52MOErA/*/i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErG/*C*G*A*C*T*A*T*A*C*G* /i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErC/*/32MOErT/ 3′ | 2′MOE/DNA, chimera, All PS ‡ |
| 5′ +G*+C*+G*C*G*A*C*T*A*T*A*C*G*+C*+G*+C 3′ | Affinity Plus/DNA chimera, All PS |
| 5′ mA*mC*mG*mC*mG*C*G*A*C*T*A*T*A*C*G*mC*mG*mC*mC*mU 3′ | 2′OMe/DNA chimera, All PS |
| 5′ /52MOErA/*/i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErG/*/iMe-dC/*G*A*/iMe-dC/*T*A*T*A*/iMe-dC/*G*/i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErC/*/32MOErT/ 3′ | Gapmer with 5-Methylcytosine |
* = Phosphorothioate bonds
/52MOEr_/, /i2MOEr_/, /32MOEr_/ = 2'MOE (2'-O-methoxyethyl base) ‡
m = 2'OMe (2'-O-methyl base)
+ = Affinity Plus (locked nucleic acid base)
/iMe-dC/ = 5-Methylcytosine (5-methyl-dC)
‡ IDT's unique 2'MOE modification
codes can be found
here.
| Chemical Modification | Common Name | Class of Modification | Activates RNase H1 | Binding Affinity Effects | Action |
|---|---|---|---|---|---|
| Phosphorothioate | PS | Backbone | Yes | Decreases | Binds non-specifically to proteins, exists as two stereoisomers |
| 2'-O-Methyl | 2'OMe | Sugar | No | Increases | Natural RNA modification; decreases toxicity |
| 2'-O-Methoxyethyl | 2'MOE | Sugar | No | Increases | Reduces protein binding, decreased toxicity |
| Locked nucleic acids | Affinity Plus™ | Sugar | No | Increases | High binding affinity at the cost of increased toxicity |
| 5-Methylcytosine | 5-MedC | Base | Yes | Increases | Natural RNA modification; prevents activation of TLR9 in CpG motifs |
Technical overview
Antisense oligonucleotides (ASOs) can be used to inhibit gene expression both in vitro and in vivo. Improvements in the design and chemistry of antisense compounds have enabled this technology to become a routine tool in basic research, genomics, target validation, and drug discovery. ASOs are increasingly used to confirm phenotypes obtained from RNAi-mediated gene silencing experiments, also.
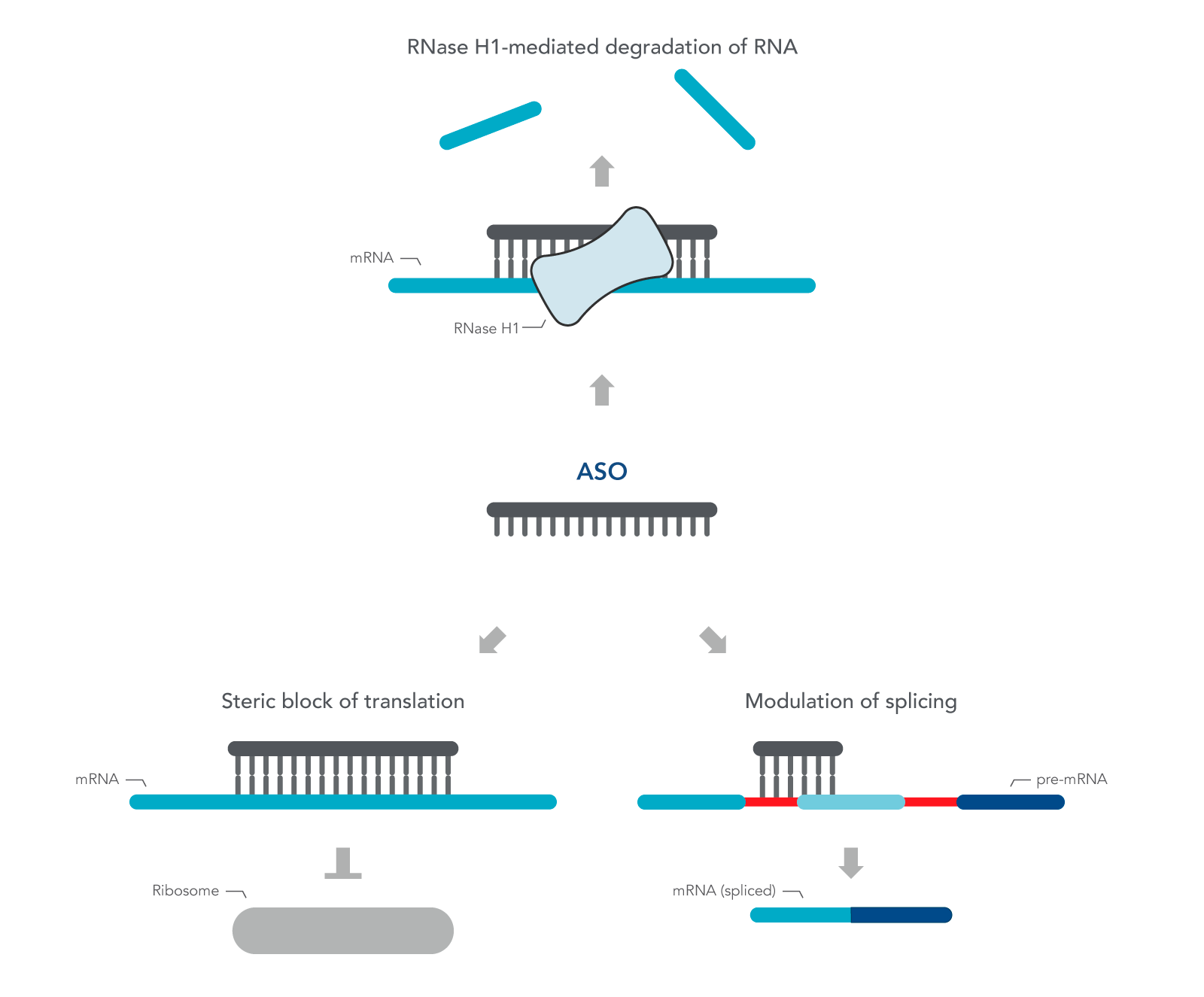
ASOs are nucleic acid sequences made as synthetic oligonucleotides, usually 15–22–bases long, containing a phosphorothioate-modified DNA segment of at least six bases. They are designed in antisense orientation to the RNA of interest, hence the name. To inhibit gene expression, they are introduced into the cell or organism, where they bind the target RNA to form an RNA/DNA heteroduplex, which is a substrate for endogenous cellular RNase H (Figure 1) [2,3]. The resulting decrease in RNA levels can be measured using RT-qPCR or RNA-seq.
ASOs can also be designed to investigate the role of alternatively spliced mRNA. Alternative splicing is one of many ways gene expression is modulated to respond to changing environmental conditions or developmental cues. ASOs are designed to the pre-mRNA sequence, and therefore, are complementary to the exon and intron junction. Unlike above, these ASOs create a double-stranded region that sterically blocks splicing factors from binding. These ASOs are designed with 2' modifications on the sugar moiety throughout the length of the sequence in order to prevent RNase H activation.
Figure 1. Antisense oligonucleotide (ASO) mechanisms of action. ASOs can modulate gene expression through RNase H1 – mediated recognition of an RNA:DNA hybrid followed by degradation of the target RNA. Alternatively, ASOs can bind to an RNA functional domain to sterically block proteins or other nucleic acids from binding at that site.
Phosphorothioates and chimeric oligos
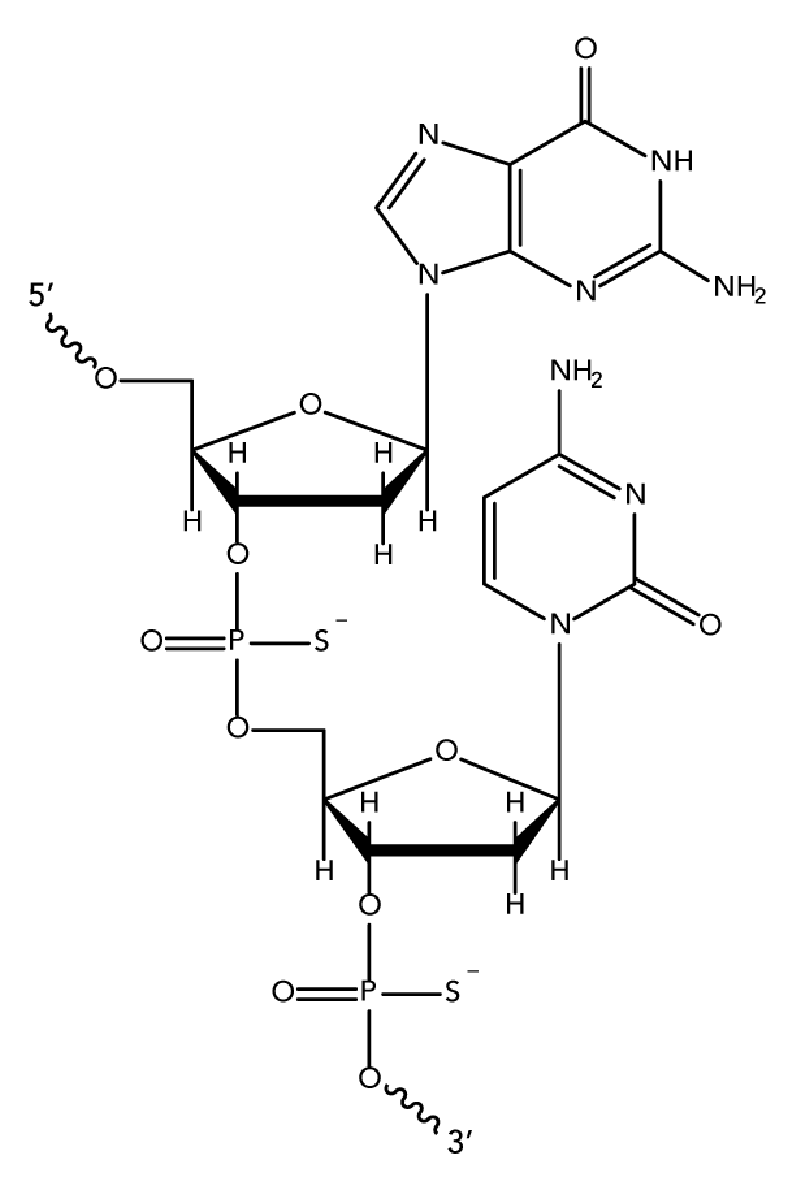
While unmodified oligodeoxynucleotides can display some antisense activity, they are subject to rapid degradation by endo- and exo-nucleases. Many 2′-O-modified RNA (such as 2′OMe RNAs and Affinity Plus locked nucleic acid bases) are sensitive to exonuclease degradation, as well. The simplest and most widely used nuclease-resistant chemistry available for antisense applications is the phosphorothioate (PS) modification. In phosphorothioates, a sulfur atom replaces a non-bridging oxygen in the oligo phosphate backbone. In the IDT ordering system, an asterisk indicates the presence of a phosphorothioate internucleoside linkage. PS oligos can show greater non-specific protein binding than unmodified phosphodiester (PO) oligos, which can cause toxicity or other artifacts when present at high concentrations.
We recommend phosphorothioate modification of ASO sequences to provide stability. Phosphorothioate linkages also promote binding to serum proteins, which increases the bioavailability of the ASO while facilitating productive cellular uptake.
Chemically modified RNA and DNA bases for chimeric antisense designs
State-of-the-art antisense design employs chimeras with both DNA and modified RNA bases [1]. The use of modified RNA, such as 2′-O-methoxy-ethyl (2′-MOE) RNA, 2′-O-methyl (2′OMe) RNA, or Affinity Plus locked nucleic acid bases in chimeric antisense designs, increases both nuclease stability and affinity (Tm) of the antisense oligo to the target RNA [4–6]. However, these modifications do not activate RNase H cleavage. The preferred antisense strategy is a "gapmer" design which incorporates 2′-O-modified RNA or Affinity Plus locked nucleic acid bases in chimeric antisense oligos that retain an RNase H activating domain. As many 2′-O-modified RNA (such as 2′OMe RNAs and Affinity Plus locked nucleic acid bases) are sensitive to exonuclease degradation, we recommend phosphorothioate modification of the ASO sequence to provide stability (See the "Phosphorothioates and chimeric oligos" section above).
It can be beneficial to substitute 5-methyl-dC for dC in the context of CpG motifs. Substitution of 5-methyl dC for dC will slightly increase the Tm of the antisense oligo. The use of 5-methyl dC in CpG motifs can also reduce the chance of adverse immune response to Toll-like receptor 9 (TLR9) in vivo. We recommend standard desalt purification for most antisense applications. For use in live animals, higher purity oligos may be required. In these instances, HPLC purification combined with Na+ salt exchange followed by end-user ethanol precipitation of the antisense oligo is recommended to mitigate toxicity from residual chemicals that may carry over during synthesis.
Product data
Antisense oligonucleotides with different chemical modifications (2'OMe, 2'MOE and Affinity Plus) exhibit different levels of inhibition of gene expression
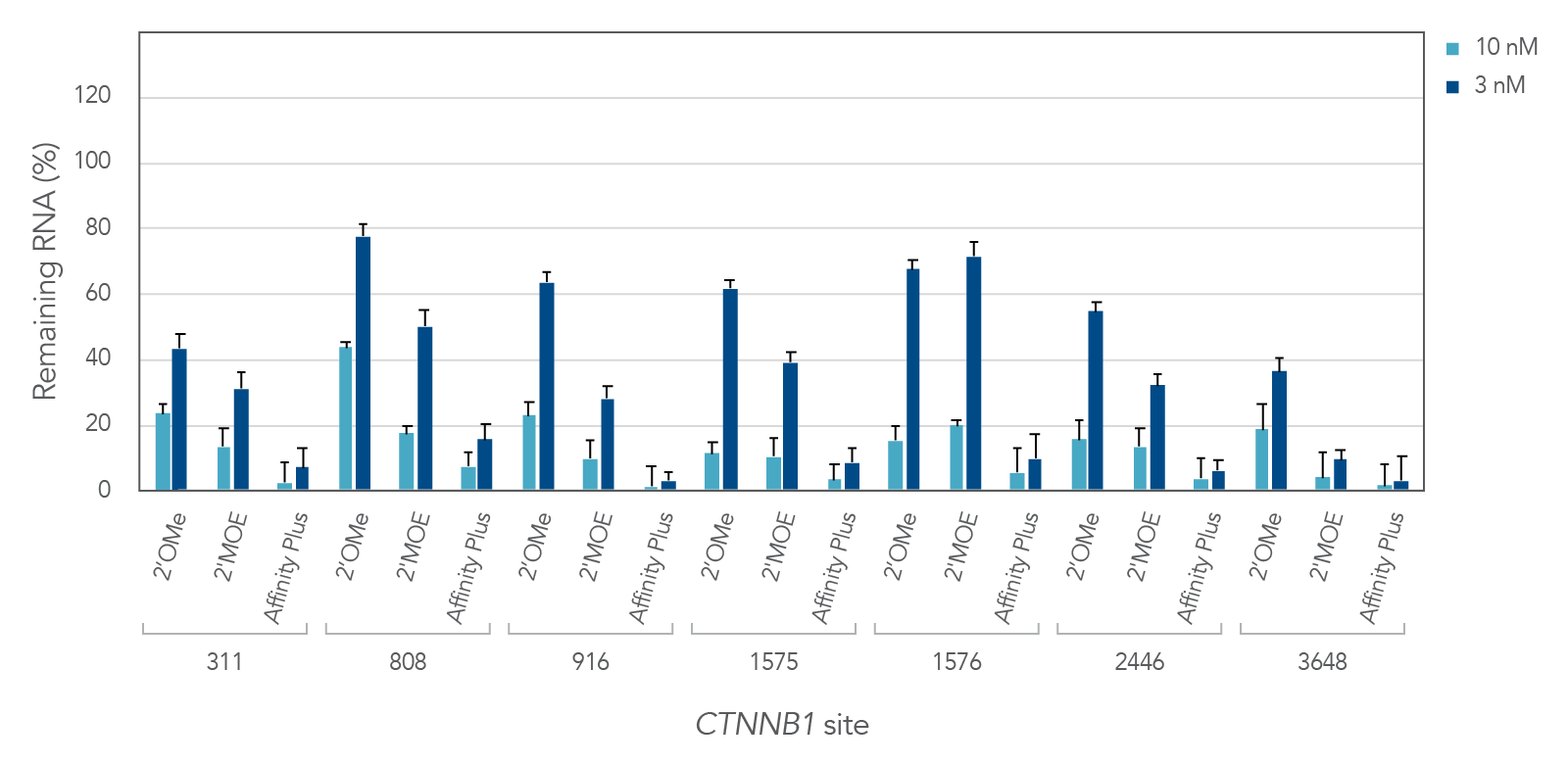
Figure 2 demonstrates the differential inhibition effect of three different modifications on seven different targeted sites of the CTNNB1 gene. Affinity Plus ASOs show the highest inhibition among the three modifications, followed by 2’MOE and 2’OME ASOs, respectively. Whereas the Affinity Plus gapmer ASOs show the highest level of inhibition, we recommend screening more sites to mitigate possible toxicity.
Figure 2: Potency comparison of 2'OMe, 2'MOE and Affinity Plus gapmer ASOs. Gapmer ASOs were designed to target multiple sites throughout the human CTNNB1 gene. The 2’OMe and 2’MOE gapmers are 20mer 5-10-5 constructs, while the Affinity Plus gapmers are 16mer 3-10-3 constructs positioned centrally within the 20mer target space (two bases are trimmed off each end of the 20mer site). ASOs were delivered in triplicate into HeLa cells with Lipofectamine™ 2000 (Invitrogen). After 24 hours, RNA levels were measured by RT-qPCR. CTNNB1 levels were calculated using the internal reference genes HPRT and SFRS9 and compared to negative control sequences with the same chemical modification composition. Error bars represent SEM, n = 3.
Resources
Frequently asked questions
What types of modifications are routinely used in antisense experiments?
Phosphorothioate (PS) bonds are added to antisense oligonucleotides to protect them from nuclease degradation. Antisense oligos can include a full PS backbone; however, the Tm decreases with each PS bond added. Including extensive PS modification can also promote the antisense oligo to non-specifically bind to proteins. This characteristic can increase oligo circulation time and improve cellular internalization when a delivery tool is not employed. However, it can also cause a pro-inflammatory response if the antisense oligo binds non-specifically to immune receptors.
5-Methyl dC (5-Me dC) modified bases are often used to prevent CpG motifs in an antisense oligo from triggering a TLR9 innate immune response. 2’-O-methoxy-ethyl (2’-MOE) or Affinity Plus locked nucleic acid base modifications are often added to increase nuclease resistance and binding affinity of antisense oligos.
For more information about the biophysical properties of the various chemical modifications used in antisense application, refer to Lennox KA, Behlke MA. Chemical Modifications in RNA Interference and CRISPR/Cas Genome Editing Reagents. Methods Mol Biol. 2020;2115:23-55.
Does IDT offer ASO design services?
Our Application Support team would be glad to help with ASO design projects. Contact us and provide your project details, including:
- Species
- Target fasta sequence or accession number
- Application (RNA knockdown, steric-blocking, splice-switching)
Do ASOs need to be HPLC purified?
Antisense oligos (ASOs) typically do not require HPLC purification due to their short length. Standard desalt purification is suitable for most antisense applications.
For use in live animals, higher purity oligos may be required. In these instances, HPLC purification combined with Na+ salt exchange followed by end-user ethanol precipitation of the antisense oligo is recommended to mitigate toxicity from residual chemicals that may carry over during synthesis.
How can I assess knockdown of my target gene?
IDT recommends using reverse transcription quantitative PCR (RT-qPCR) to assess knockdown of your RNA target. The IDT PrimeTime™ Predesigned qPCR Primer and Probe Sets or a custom PrimeTime qPCR Assay can be used to assess gene expression for human, mouse, and rat transcriptomes.
We recommend at least two qPCR assays designed a distance apart from each other on the transcript to help control for artifacts. For example, you may get a false negative result if there are gene isoforms that are not targeted by the ASO but are still quantified by the qPCR reaction. Alternatively, you may get a false positive result if the ASO blocks the reverse transcriptase enzyme during cDNA synthesis, thus preventing full-length RNA transcript conversion to cDNA. Having concordant data for multiple qPCR assays can help increase confidence in your results.
How much ASO should I use in my knockdown experiment?
Optimal ASO concentration is dependent on both the cell type or organism as well as the delivery method.
For delivery of Antisense Oligos (ASOs) to immortalized tissue culture cell lines using cationic lipids, IDT typically uses an ASO dose response in the 1–30 nM range. Optimization may be needed for your specific target and system.
What is the purpose of ASOs modified in a gapmer format?
Gapmer antisense oligonucleotides (ASOs) include modified bases at each end of the Antisense Oligo (such as 2’MOE or Affinity Plus modified bases) with an internal stretch of unmodified DNA bases. The terminal modified bases can help to enhance stability and increase binding affinity to the target. The internal unmodified DNA bases are necessary to enable RNase H to bind to and cleave the RNA target.
What is the difference between 2’MOE and Affinity Plus™-modified ASOs?
2’MOE and Affinity Plus modified bases are often included in ASOs, in addition to phosphorothioate bonds. 2’MOE modified bases help to increase nuclease resistance and binding affinity and decrease toxicity of the ASO. Affinity Plus bases are locked nucleic acid bases that also help to increase binding affinity and nuclease resistance but can be toxic.
2’MOE bases are often incorporated into ASOs in a 5-10-5 gapmer format, with five 2’MOE bases, ten DNA bases, and five 2’MOE bases. Affinity Plus bases are typically used in shorter ASOs in a 3-10-3 gapmer format (three Affinity Plus bases, ten DNA bases, three Affinity Plus bases). The shorter length of Affinity Plus ASOs is beneficial when delivering the ASO in vivo without a delivery tool.
Which is the best way to deliver oligos into cells in culture?
The best method depends on the type of cells and types of modifications on the oligo, as well as the intended use of the ASO. Contact us for more information.
How do I order a phosphorothioate modified oligo?
IDT uses a simple notation for phosphorothioate bonds (PS bonds). Insert an asterisk (*) between the bases within your sequence to denote a phosphorothioate chemical modification.
The following example has a PS bond between the T and G bases: 5'- TTTTTT*GGGGGG-3'. PS bonds introduced between the last nucleotides at the 5'- or 3'-end of the oligo inhibit exonuclease degradation.
Order PS-modified oligos at: idtdna.com/site/order/oligoentry.
How do I order 2'-O-Methyl modified bases?
2'-O-Methyl bases are ordered under "Custom DNA Oligos." Represent the 2'-O-Methyl bases in your sequence with a lowercase "m" in front of each base.
If the sequence contains unmodified RNA bases the sequence would need to be ordered as a "Custom RNA Oligo" using the same notation for 2'-O-Methyl bases.
Can a phosphorothioated oligo for antisense studies be used in medium containing serum, and what will be its half-life?
Antisense oligos containing phosphorothioate bonds can be used in medium containing serum. However, it is recommended that you inactivate the exonucleases in the serum to decrease the chances of degradation even when protecting modifications (such as phosphorothioate bonds) are present. Exonuclease inactivation can be accomplished by heating at 65°C (instead of 56°C) for 30 minutes.The half life of such an antisense oligo will be dependent on the target transcript, the cell type, and the antisense construct, resulting in no set average for this number.
Examples of oligonucleotide stability studies in serum can be found in the following references:
- Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27(9):1788-1799.
- Lennox KA, Owczarzy R, Thomas DM, Walder JA, Behlke MA. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Mol Ther Nucleic Acids. 2013;2(8):e117. Published 2013 Aug 27.
References
- Lennox KA, Behlke, MA. Mini-review on current strategies to knockdown long non-coding RNAs. J Rare Dis Res Treat 2016; 1 (3): 66–70.
- Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA. 1988;85(14):5011–5015. doi:10.1073/pnas.85.14.5011
- Dagle JM, Walder JA, Weeks DL. Targeted degradation of mRNA in Xenopus oocytes and embryos directed by modified oligonucleotides: studies of An2 and cyclin in embryogenesis. Nucleic Acids Res.1990;18(16): 4751–4757. doi:10.1093/nar/18.16.4751
- Braasch DA, Liu Y, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30(23):5160–5167. doi:10.1093/nar/gkf651
- Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligos stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30(9):1911-1918. doi:10.1093/nar/30.9.1911
- Grünweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2'-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 2003;31(12):3185-3193. doi:10.1093/nar/gkg409