xGen™ SARS-CoV-2 Hybridization Panel
Strain-agnostic enrichment of SARS-Cov-2 genome
The xGen SARS-CoV-2 Hyb Panel spans the complete viral genome and is not susceptible to known and novel mutations arising from diverse strains.
xGen NGS—made for SARS-CoV-2 research.
Ordering
- Identify known and novel mutations arising from diverse strains
- Available as a bundle with xGen™ Universal Blockers-TS Mix and xGen Hybridization and Wash Kit
For the xGen Hybridization and Wash Kit, xGen Universal Blockers, xGen Library Amplification Primer Mix, and/or xGen Human Cot DNA, please visit the xGen Hybridization Capture Core Reagents page.
Product data
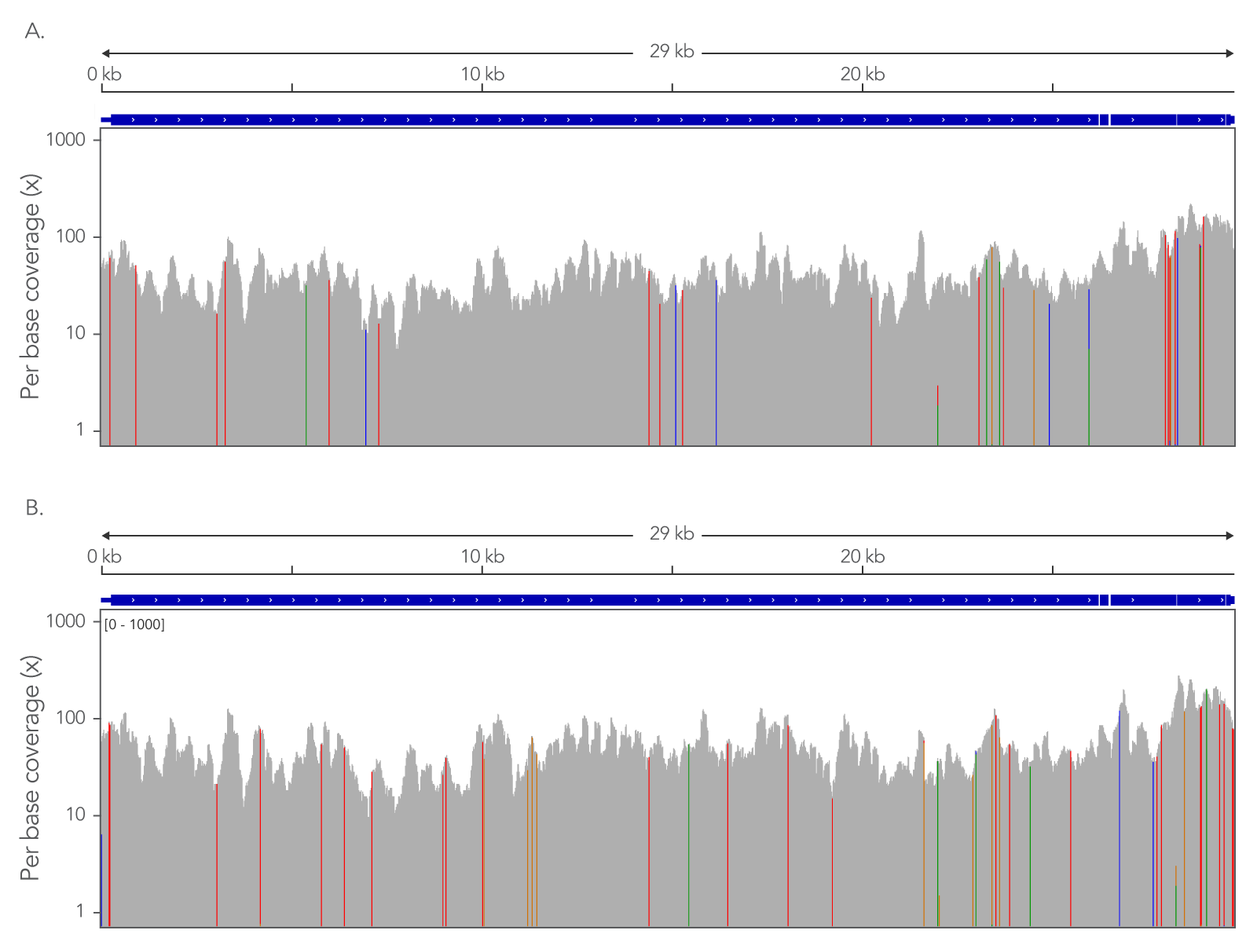
Even coverage across the complete viral genome
Figure 1. An example of the xGen™ SARS-CoV-2 Hyb Panel providing even coverage across the complete viral genome allowing strain identification with minimal read depth. (A) Coverage of a nasopharyngeal swap sample identified as belonging to strain Alpha (B.1.1.7). (B) Coverage of a nasopharyngeal swap sample identified as belonging to strain Delta (B.1.617.2). 5 µL of total RNA sample (previously shown to contain viral sequence via qPCR assay) were used to prepare NGS libraries (n=7) using the xGen™ Broad Range Library Prep Kit. Libraries were quantified and equal amounts were captured with the xGen™ Hybridization and Wash Kit. Samples were sequenced on an Illumina MiSeq™ and subsampled to 25000 total reads per sample. Data was aligned to the Refseq sequence (NC_045512) and viewed in the Integrated Genome Viewer (IGV). The IGV view color codes bases that do not match the canonical sequence. Strains were identified by submitting a consensus sequence for the sample to https://pangolin.cog-uk.io/