Locked nucleic acids and their applications
Overview
Increase oligonucleotide hybridization melt temperature (Tm), target affinity, nuclease resistance, and mismatch discrimination.
What are locked nucleic acids?
Locked nucleic acids are modified RNA monomers. The “locked” part of their name comes from a methylene bridge bond linking the 2′ oxygen to the 4′ carbon of the RNA pentose ring (Figure 1). The bridge bond fixes the pentose ring in the 3′-endo conformation.
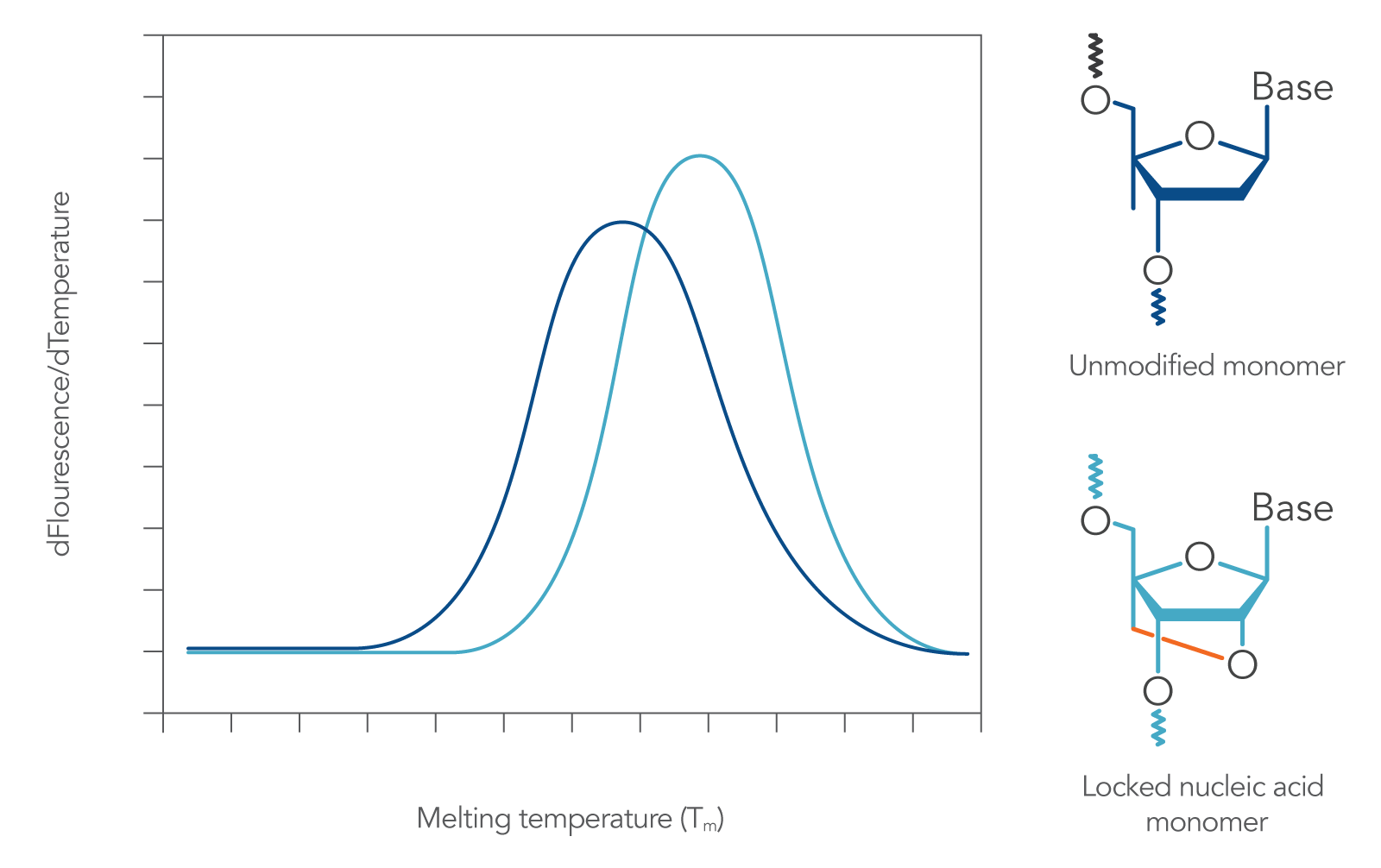
These bases follow Watson-Crick base-pairing rules when mixed with DNA or RNA bases in an oligonucleotide. When incorporated into a probe, they impart heightened structural stability, leading to increased hybridization Tm (Figure 2)[1]. Locked nucleic acids also provide resistance to nucleases, enzymes that would otherwise degrade the DNA or RNA sequence.
Figure 2. Incorporation of locked nucleic acid bases increases sequence melting temperature. When locked nucleic acid modified bases are incorporated into a DNA sequence (such as a qPCR probe), its duplex melting characteristics are changed, resulting in increased Tm [2].
Manage sequence Tm using locked nucleic acid bases
Because of the afforded increase in Tm, locked nucleic acid qPCR probes can be designed with shorter lengths than standard probes [2]. Shorter probes have better quenching and a higher signal-to-noise ratio than unmodified longer probes [2].
The locked nucleic acid bases included in these sequences enhances discrimination of thermodynamically similar samples such as single nucleotide polymorphisms and transcript variants. This improves their ability to distinguish mutations or single nucleotide polymorphisms (SNPs)[3]. By including locked nucleic acid bases, you can design a probe with up to a ΔTm >15°C. By varying the number of these modified bases, you can essentially control the Tm of a nucleotide duplex. Use these probes in methods that use differential hybridization to distinguish polymorphisms.
Other applications of locked nucleic acid probes include the identification of transcript variants, microbial species, as well as of targets in FFPE tissue, biofluids, and other challenging samples.
Locked nucleic acid oligonucleotides give control over hybridization
Locked nucleic acid oligonucleotides are useful in template switching oligo (TSO) designs and for strengthening target oligo binding in challenging sequence regions [4]. As with locked nucleic acid qPCR probes, the hybridization Tm can be manipulated by the number of locked nucleic acid bases incorporated.
Improve your hybridization applications with locked nucleic acid technology
IDT provides 2 types of locked nucleic acid products: custom Affinity Plus™ DNA & RNA Oligonucleotides and Affinity Plus qPCR Probes. Affinity Plus qPCR Probes and custom Affinity Plus DNA & RNA Oligonucleotides provide identical results to traditional locked nucleic acid sequences and offer a cost-effective approach to applications that call for locked nucleic acids.
Related products
Resources
References
- You Y, Moreira BG, Behlke MA, et al. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34(8):e60.
- Ugozzoli LA, Latorra D, Puckett R, et al. Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal Biochem. 2004;324(1):143-152.
- Mouritzen P, Nielsen AT, Pfundheller HM, et al. Single nucleotide polymorphism genotyping using locked nucleic acid (LNA). Exper Rev Mol Diagn. 2003;3(1):27-38.
- Harbers M, Kato S, de Hoon M, et al. Comparison of RNA- or LNA-hybrid oligonucleotides in template-switching reactions for high-speed sequencing library preparation. BMC Genomics. 2013;14:665.