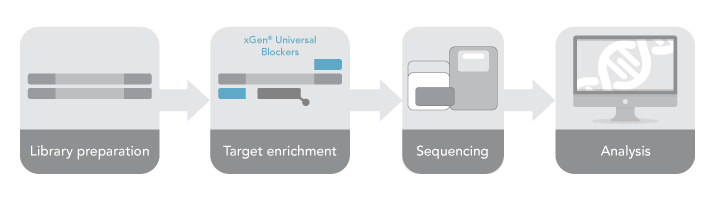
Understanding more precisely the nuances of performing target capture translates directly into obtaining the most clarity from your data during downstream analysis. To complement the evolving next generation sequencing (NGS) workflow (Figure 1) and suite of target enrichment products IDT offers, here our research scientists provide specific guidelines to achieve reliable, repeatable results with our xGen™ Hybridization Capture protocol (xGen Hybridization Capture of DNA libraries).
Tips to improve NGS target capture
- Carefully select a hybridization capture panel for your samples and targets. IDT offers the xGen Hybridization Capture Panels, xGen Blocking Oligos, NGS adapters, xGen Hybridization and Wash Kit, and the xGen Library Amplification Primer Mix—to help you get the most out of your hybridization workflow. For custom probes, use the IDT Target Capture Probe Design & Ordering Tool. The xGen Hybrid Capture system was specifically designed to deliver reliable and powerful NGS data.
- If your experiment focuses on non-human library captures, keep in mind that Human Cot DNA might not be ideal. Instead, we recommend alternatives like mouse Cot DNA, or Salmon sperm DNA.
- Check that lab instruments used in the protocol have been recently calibrated. Even small changes in temperature (+/- 2°C) for hybridization reactions and washes have an impact on the flanked-on target percentage and GC bias.
Skewed GC bias in NGS data is a known indicator for poor capture uniformity, thus calibration is an important consideration. A hotter wash temperature leads to drop-out of low GC regions from capture. A colder wash temperature leads to a lower
on-target percentage.
Note: For target enrichment sequencing it is important to quantify how many of your sequencing reads were on-target verses off-target. The simplest approach is to calculate the number of aligned bases on your target region. However, this can be complicated to apply across libraries with different insert lengths. For example, if one of your targets is a 150 bp read and the insert is 150 bp, then your on-target bases would be 100%. Meanwhile, a larger 300 bp read, aligned to the same 150 bp target, would mean that only 50% of the bases are on-target. To simplify the analysis, and compare across diverse samples, the target region is usually padded (150 bp in either direction) to calculate bases flanked on-target (Figure 2).
- Use a plate protocol when processing multiple samples. Using the plate protocol (versus the tube protocol) shows lower sample-to-sample variability within one experiment.
- When working with plates, avoid using the wells on the perimeter of the plate’s edges. Evaporation is more likely to occur in these outer rows.
- Always ensure that the hybridization reaction tube or plate is tightly sealed.
Evaporation of the hybridization reaction can lead to capture failure.
Note: Optimization of the sealing method might be necessary if heat sealers are used.
- Extending hybridization incubation time from 4 to 16 hours may improve panel performance, particularly for smaller panels (< 1,000 probes).
- Preheat the “heated wash buffers” a minimum of 15 minutes before use. Preheating helps ensure there is sufficient time to heat the buffers to 65°C at critical steps during the protocol.
- When using the optional Appendix A: AMPure XP Bead DNA concentration protocol, make sure that no SPRI beads are accidentally carried over into the hybridization reaction. Bead carryover has a negative impact on flanked on-target percentage and probe coverage.
- Understand that the optional Appendix A: AMPure XP Bead DNA concentration protocol requires more Human Cot DNA than the standard SpeedVac method. Additional Human Cot DNA (IDT Cat# 1080768; 1080769) must be purchased separately from the xGen Hybridization and Wash Kit. Using a standard amount of Human Cot DNA with the AMPure XP Bead DNA concentration protocol can lead to a lower flanked on-target percentage due to a loss of small human Cot DNA fragments (50–300 bp fragment size) during AMPure XP concentration.
- Vortex every 10 to 12 minutes during the 45-minute bead capture to improve the kinetics of the capture.
- Do not let the Streptavidin beads dry out at any point during the protocol. If necessary, extend washes slightly rather than let the beads dry out.
- Ensure the Streptavidin beads remain fully resuspended during room temperature washes. Vortex vigorously and adhere to the incubation guidelines during room temperature washes to improve data quality.
Note: Bead splashing on the plate seal has no negative impact on capture results.
- Always use fresh seals in each step of the protocol that calls for adhesive seals on the plates. Using fresh seals avoids possible sample cross-contamination during the plate protocol.
More tips for hybridization capture
The xGen hybridization capture of DNA Libraries protocol specifies the guidelines and required steps to follow for target enrichment of a library prepared from genomic DNA. We recommend using the accompanying xGen Predesigned Hyb Panels or xGen Custom Hyb Panels and to use the xGen Universal Blockers to reduce nonspecific binding of adapter arms. Let these reliable products and protocol guidelines equip you for success in your NGS target enrichment applications.
Learn more about the xGen family of hybridization capture products.
RUO22-1180_001.1